Week 2: Python basics
Taken/adapted from: https://ucdavis-bioinformatics-training.github.io/2022-Feb-Introduction-To-Python-For-Bioinformatics/python/python2
Objectives
- Introduce you to as many concepts in python as possible.
- Python’s basic data and data structures
Setting up
Make sure you have VSCode and python installed. Let’s write our first piece of code.
-
Open a new file in VSCode.
- Copy and paste (or type) this into your new file:
print("Hello, World!") -
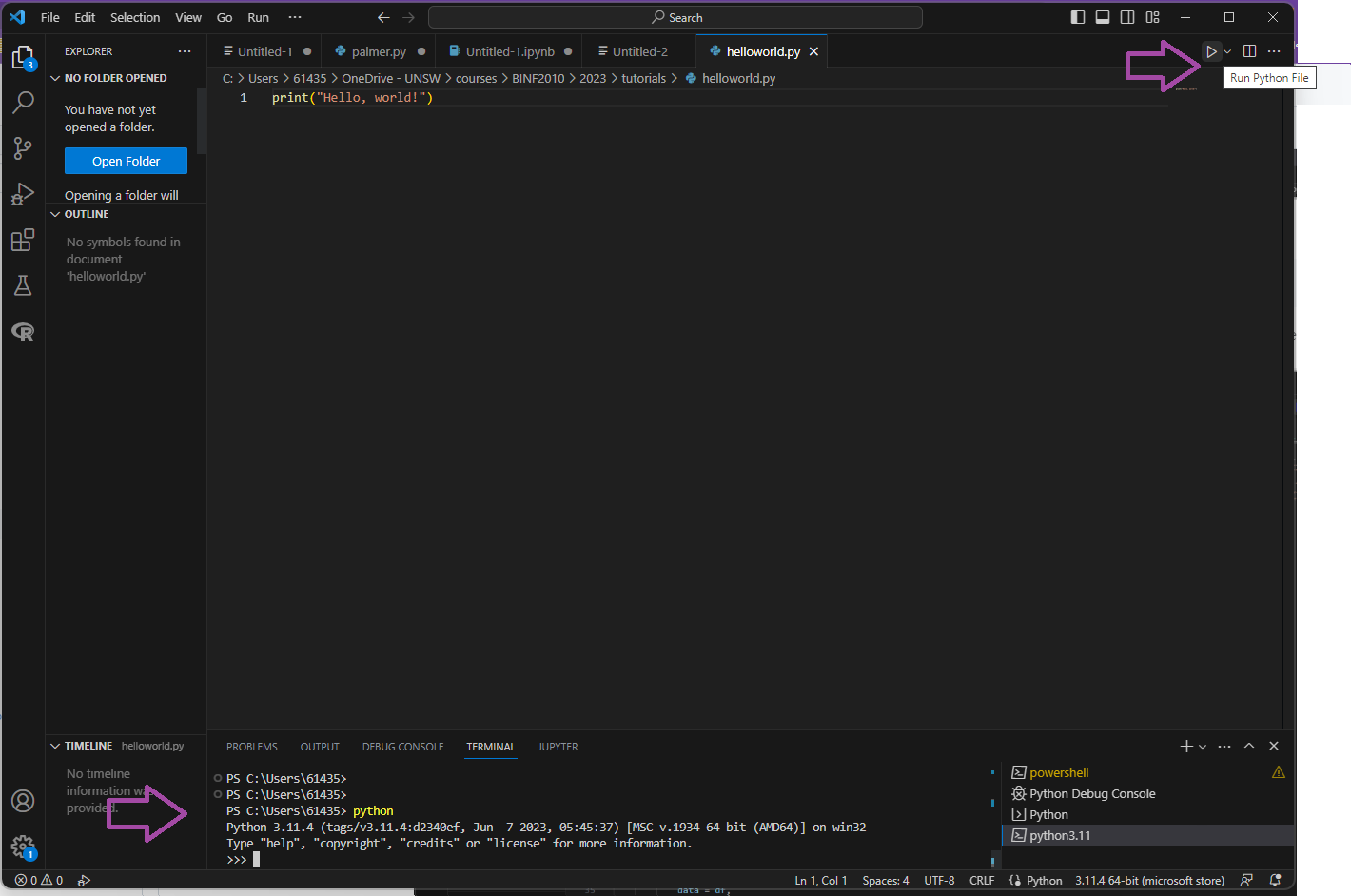
Save the file and name it “helloworld.py”. Note the “.py” extension. Click the triangle “play” button to run your code in the terminal. Python can also be run interactively. Type in “python” into the terminal. Paste/type in the code snippets in the terminal.
-
Finally, you can also run python in a Jupyter notebook. The extension for this is “.ipynb”. Each “chunk” of code can be run within the notebook.
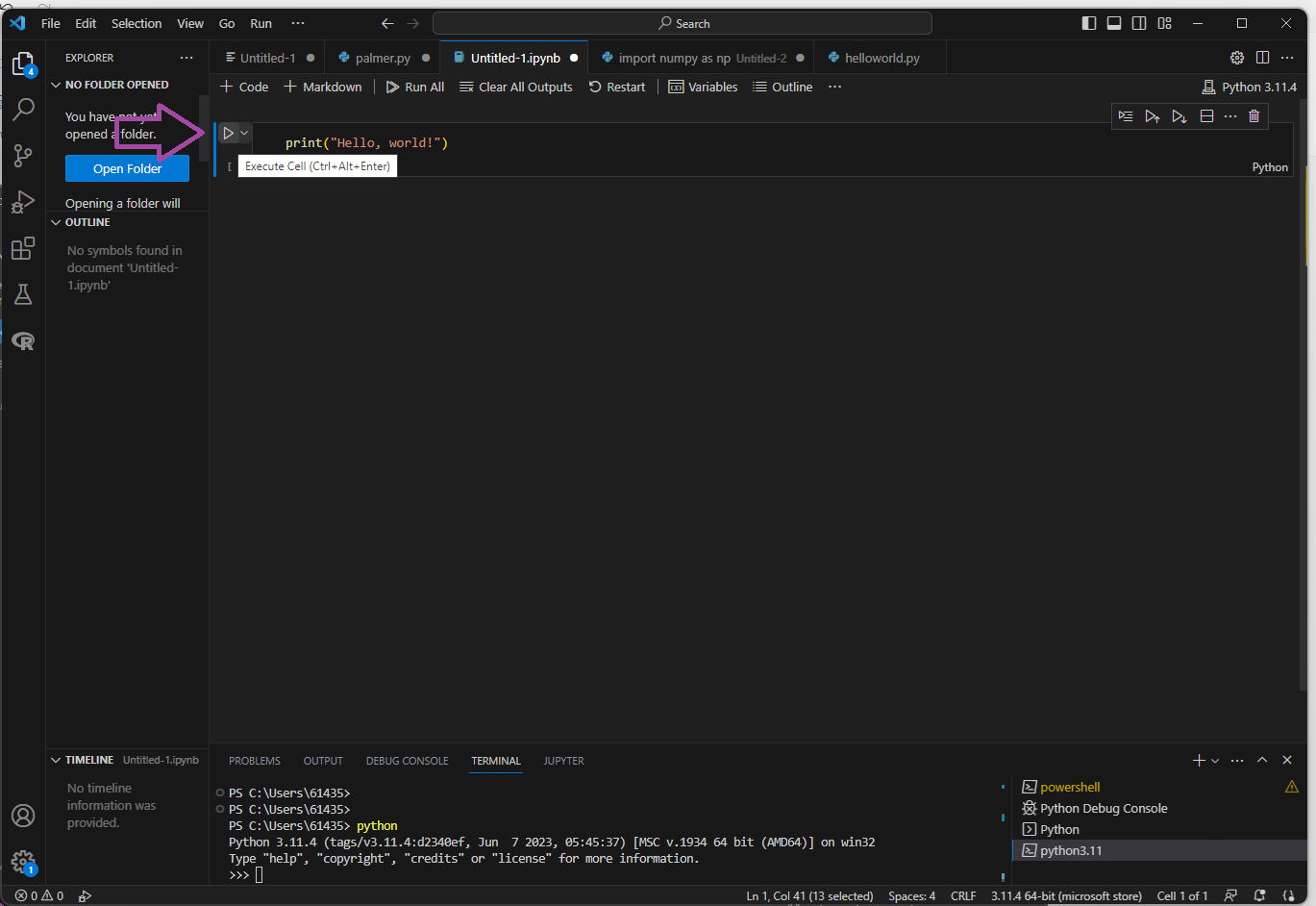
- Save a Jupyter notebook as “yourname_week2.ipynb”. You can write and run your code chunks in this file, and make sure to save! For the remainder of this tutorial, you will be copying bits of code and pasting it there.
Definitions
- Variables: way to store information that can be named, accessed, and changed in a computer program
- Data types: the type of values a variable can have, and also what mathematical, relational or logical operations can be applied to it (without causing an error!)
- Casting: a way to convert values for certain data types to other data types
Variables
- “Named” storage.
- Can store numbers, characters, etc.
- Variable names can only contain alphanumeric characters (A-Z, a-z, 0-9) and the underscore (_). No other special characters can be used, and cannot start with a number.
- Type is determined by the value assigned
- Assign values with the equals sign =
Basic Data Types:
Built-in datatypes and their corresponding category:
- Text Type: str
- Numeric Types: int, float, complex
- Sequence Types: list, tuple, range
- Mapping Type: dict
- Set Types: set, frozenset
- Boolean Type: bool
- Binary Types: bytes, bytearray, memoryview
- Examples of the most common data types:
int: integers, these are whole numbers, negative or positive.
n = 42
float: floats, or real numbers (i.e., doubles)
n_pi = 3.14
- str: strings, any text (including all symbols and spaces), designated by quotes.
gene = "TAF1"
print(gene)
print("My gene is called:", gene)
print("My gene is called:" + gene)
Note, print outputs to your screen. What is the difference in output between the last two print statements?
- bool: booleans, true/false. Represents the two values of logic and Boolean algebra
control = False
treatment = True
- The
type()function returns the data type of the variable
print("The data type of the variable 'n' is:")
print(type(n))
print("The data type of the variable 'gene' is:")
print(type(gene))
Built-in string methods
Strings have a long list of built-in methods to return modified versions of the string.
- The methods do not modify the string, they return a new string. Test this out:
tmpstr = "Hello my name is X"
allcaps = tmpstr.upper()
print(allcaps)
newstr = tmpstr.replace("X","Y")
print(newstr)
- Concatenate strings: + symbol
tmpstr2 = "How are you doing?"
print(tmpstr + " " + tmpstr2)
- Find the position of one string in another. Note, returns -1 if not found.
print(tmpstr2.find("you"))
print(tmpstr2.find("california"))
More here: https://www.w3schools.com/python/python_ref_string.asp
Q1: Test yourself!
In your jupyter notebook, create new chunk for this question. In this chunk, write some code where you assign your name to a variable called
user. Then print out a sentence that reads “This is my NAME codebook”, where NAME is replaced by yourusertext.
Casting
Casting is converting certain values for certain datatypes to other datatypes. Some examples are listed below.
- Convert a string to a boolean with
bool()
tmpstr = "Hello"
tmpbool = bool(tmpstr)
print(tmpbool)
- Convert a decimal to a string with
str()
n = 42.24
print("The number is: " + str(x))
- Convert string to an integer with
int()
mystr = "50"
myint = int(mystr) + 1
print(str(myint))
Mathematical, relational and logical operations
Comparisons
Comparisons are useful and some of them most common operations performed. Numerical and string comparisons can be done like so:
- Equalities (==)
- Inequalities (>,<, >=, <=)
print(1<1)
print(1<2)
print(2>1)
print(1<=1)
print(2>=1)
print(1==1)
print(0==1)
gene = "TAF1"
greeting = "hello"
print(gene == "BRCA2")
print(greeting == "hello")
Arithmetic
- Addition
a = 42
b = 7
print(a + b)
- Subtraction
print(b - a)
- Add/subtract and assign
c = 83
c += 5
print(c)
c -= 10
print(c)
- Division
print(a/b)
- Exponents
print(4**b)
#or
expb = pow(4,b)
print(expb)
- Remainder
print(42 % 4)
- Absolute value
av = abs(24-42)
print(av)
- Round, Floor, Ceiling
print(round(4.2))
print(int(4.2))
import math
print(math.ceil(4.2))
print(math.floor(4.8))
print(int(4.8))
- The math package has many common math functions you can use: https://docs.python.org/3/library/math.html
Q2: Test yourself!
In your jupyter notebook, create new chunk for this question. In this chunk, start off by importing the “random” python package (i.e.,
import random). Then, write some code that assigns a random number to two variables (x and y) with therandom.random()function. Then calculate the average value of these variables (x and y), and round the result to the nearest two decimal points. Assign that result to the variable z, and print it out.
Basic Data Structures
In addition to simple data types, we have collections of these data types into things we call data structures. These include:
- tuples: list of elements, are immutable (cannot be modified)
- lists: list of elements, can be nested, contain different data types, and mutable (can change)
- dictionaries: a set of key/value pairs where the keys are unique (hash table)
- sets: list of UNIQUE (and immutable) elements, mutable
Tuples
- List of elements, but cannot be changed and defined by “()”.
- Functions often pass tuples (not lists) back to the user.
gene_tuple = ("DDX11L1","WASH7P","MIR6859-1","MIR1302-2HG","MIR1302-2","FAM138A")
Lists
- List of variables/elements, defined by “[]”.
gene_list = ["DDX11L1","WASH7P","MIR6859-1","MIR1302-2HG","MIR1302-2","FAM138A"]
In python, lists are 0-indexed. This means to access the first element, we use the index value of 0. However, we can also use “negative indices” to access this (based on the length of the list):
- First element
print(gene_list[0])
print(gene_list[-6])
- Last element
print(gene_list[5])
print(gene_list[-1])
- Extracting a range of the list - not very intuitive, bust uses the colon (:) to pick a range of values.
print(gene_list[-3:]) # last three elements
print(gene_list[1:3]) # elements 2 to 3
print(gene_list[:3]) # up to element 3
print(gene_list[1:2]) # second element, but returns a list
print(gene_list[1]) # second element, but returns a string
- Note, the same range can be applied to strings, where you extract ‘characters’
mystring = "The quick brown fox jumps over the lazy dog"
print(mystring[4:9])
- Length of a list
print("The length of gene_list is " + str(len(gene_list)))
- Lists can have elements of any type
forty_twos = ["42", 42, "forty-two", 42.0]
val = forty_twos[1]
print(val)
print(type(val))
val = forty_twos[2]
print(val)
print(type(val))
- Creating a new variable equal to a list does NOT create a copy, both variables point to the same list
gene_list2 = gene_list
gene_list2[2] = "DMR3"
print(gene_list)
- Copy method to make a actual copy of a list
gene_list2 = gene_list.copy()
gene_list2[2] = "DMR5"
print(gene_list)
print(gene_list2)
- Join lists
gene_list + gene_list2 - Check for membership in a list with “in”
print("BRCA2" in gene_list)
Built-in list methods
Some basic list methods that are useful include .append(), .remove(), and .reverse(). More methods can be found here: https://www.tutorialsteacher.com/python/list-methods
gene_list.append("BRCA2")
print(gene_list)
gene_list.remove("WASH7P")
print(gene_list)
gene_list.reverse()
print(gene_list)
Q3: Test yourself!
In your jupyter notebook, create new chunk for this question. In this chunk, copy the
gene_listvariable from the previous code to a new variable calledgene_list_test. Add three random genes and remove the third gene in the list. Add a gene to the middle of the list.
Dictionaries
- A set of key/value pairs where the keys are unique (hash table)
- Dictionary values are pointed to by the keys.
- Values can be anything from int, float, and bool to lists, tuples, and other dictionaries.
gene_exp_dict = {"DDX11L1":43.2,"WASH7P":45,"MIR6859-1":60.1,"MIR1302-2HG":12,"MIR1302-2":0.5,"FAM138A":23}
- Accessing a value in the dictionary by referencing the key
print(gene_exp_dict["WASH7P"])
- Overwriting a value
gene_exp_dict["WASH7P"] = 39
print(gene_exp_dict["WASH7P"])
- Adding a new value
gene_exp_dict["BRCA2"] = 100
print(gene_exp_dict)
Dictionary built-in methods
- Returns all the keys
print(gene_exp_dict.keys())
- Returns all the values
print(gene_exp_dict.values())
- Returns all the key,value pairs
print(gene_exp_dict.items())
- Check if a key exists in a dictionary
print("BRCA2" in gene_exp_dict)
- Make a copy of a dictionary
gene_exp_dict_copy = gene_exp_dict.copy()
Sets
- Collections of immutable unique elements
- These are useful to perform some operations, which can be applied faster than on lists
- To create a set, we can use either curly brackets ({}) or the set() constructor.
s1 = {1, 2, 3}
s2 = set([1, 2, 3, 4])
print(f"Set s1: {s1}")
print(f"Set s2: {s2}")
- A useful operation is creating a union of sets (merging two sets together). If two sets have two or more identical values, the resulting set will contain only one of these values.
set_union = s1.union(s2)
print(set_union)
set_union = (s1 | s2)
print(set_union)
More here: https://www.dataquest.io/blog/data-structures-in-python/
Q4: Test yourself!
In your jupyter notebook, create new chunk for this question. Create a dictionary for gene expression of multiple genes for multiple samples. Call it
gene_exp_dict. Populate the dictionary with at least 10 genes for 5 samples. Then, write the code to get the gene expression for the 4th gene, 2nd sample.
[Solutions - released next week!]
Back to the homepage